Two software programs, tcaSIM and tcaCALC detailed below, are available for download for free. To download the software, please contact Jeffry Alger, PhD here. The software is currently is beta status. Users are encourage to report problems bugs directly to us.
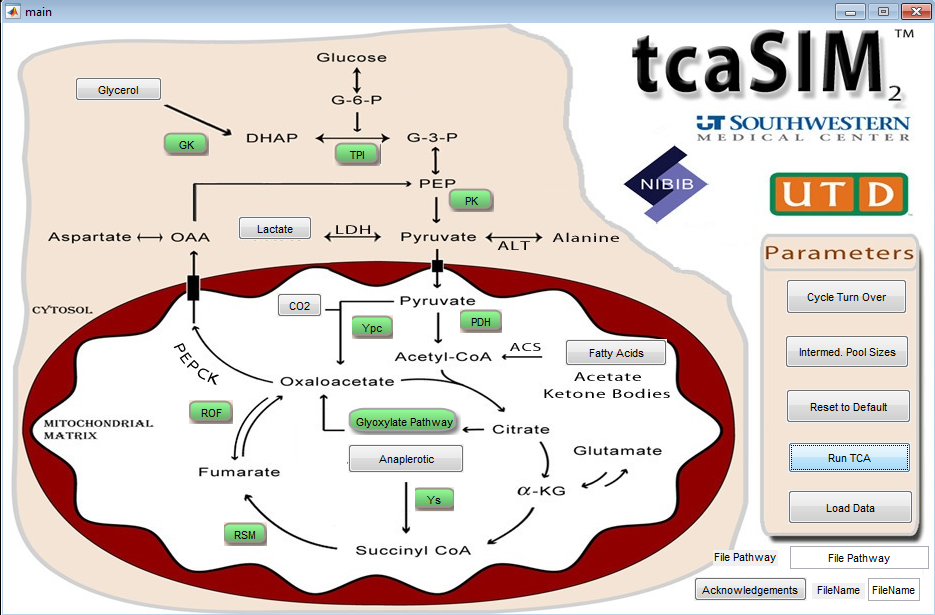
tcaSIM
The tcaSIM program is a tool for use with labeling studies in intermediary metabolism, primarily those applying 13C NMR spectroscopy, but also for 14C radiolabel studies and 13C mass isotopomer analysis. The simulation has been developed to assist in the design of experiments and to help the user understand the effect of various metabolic states and substrate enrichment levels on labeling patterns.
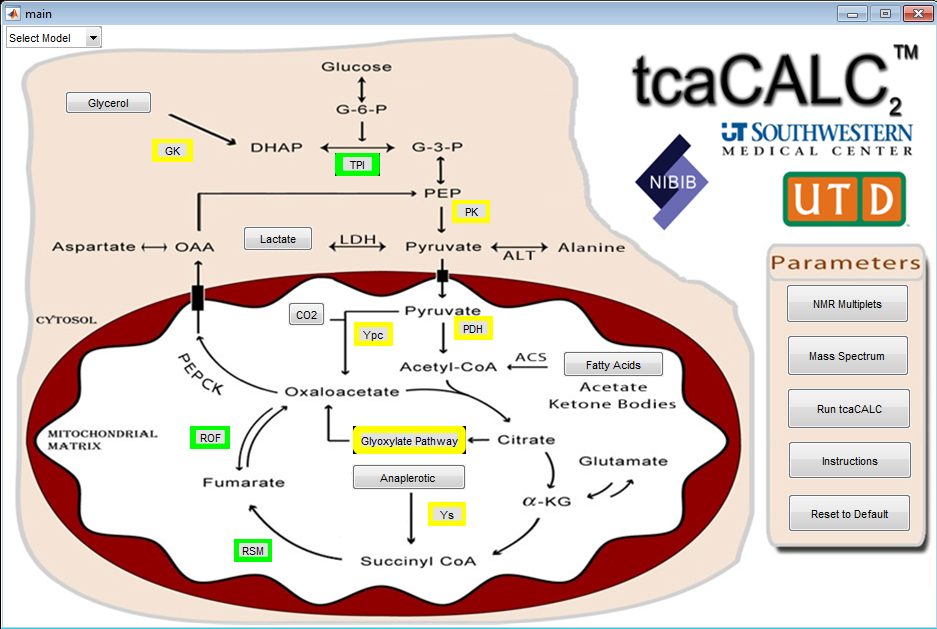
tcaCALC
The tcaCALC program performs an isotopomer analysis to estimate relative pathway fluxes in biological systems at metabolic and isotopic steady state. It is used in 13C labeling studies where biological systems (isolated cells, perfused organs or intact animals) are provided enriched substrate(s), and the labeling of intermediates detected using 13C NMR spectroscopy, mass- and tandem mass spectrometry. The 13C spectra are collected either directly from intact tissue, such as in the case of perfusions conducted inside a magnet, or after extraction of a tissue sample (isolated cell studies or tissue samples taken after in vivo infusion of 13C-enriched substrates).
Please contact Jeffry Alger directly for any information about the current versions of tcaCALC and tcaSIM.
Coming soon
Coming soon
| Date | Version | Comments |
|---|